Commentary: In silico identification of potential key regulatory factors in smoking-induced lung cancer
Salem A. El-aarag1, Mahmoud ElHefnawi2,3*
Due to the complexity and heterogeneity of cancer, it is inevitable to investigate carcinogenesis from the systems perspective1,2. This was the driver of our study3. In the next sections, we will highlight some aspects regarding to the methodology of the study and the most important findings.
In our study3, we concentrated only on the drivers of lung cancer among smokers, only taking into consideration smoking as the sole factor for lung cancer development. Smoking is a major contributor to lung cancer development4. Spira et al. (2007) made an unique experiment for studying smoking-induced lung cancer. We found that Wang and Chen (2011) analyzed only Spira’s dataset, integrated microarray gene expression profiles and information on protein-protein interactions (PPIs) and developed a network-based biomarker which established a set of 40 biomarkers with potentially important roles in lung carcinogenesis2.
Our approach included network-based and enrichment analysis of differentially expressed genes (DEGs) between normal and cancerous lung. We identified differentially expressed genes (DEGs) between normal and cancerous lung using two software (GEO2R and Python script analysis tools) with different style of outputs. The former produced single list ordered by significance, whereas the last one produced two lists (up- and downregulated genes). As Enrichment analysis is sensitive to input list size, DEGs were divided into 9 different lists according to their length and source. We concentrated on the enriched items with higher overlap among these lists so that our conclusion could be more reliable5,6.
While using atBioNet (network analysis tool), we found that using the stringent options produced good repeatability provided that using large DEG lists (n => 500). We used stringent options to secure the efficiency. We set atBioNet to add only nodes which directly connected to at least two input nodes using (K2 Human Subset Database) which is a smaller and more robust database so that the added nodes represent the most accurate hidden information7.
For enrichment analysis (Enrichr), we didn’t use the standard Fisher’s exact test as usual, but we applied the z-score method which outperformed it8.
Both approaches showed that MAPK signaling is the most significant pathway related to lung cancer in smokers. Both approaches identified that MAPK, Toll-like receptor signaling pathway, and renal cell carcinoma signaling pathways as being important in smoking-induced lung cancer (Fig. 1)9-10. Targeting MET which has been associated with both sporadic and inherited forms of human papillary renal carcinomas may be therapeutic target for treatment of a gefitinib/erlotinib-resistant lung tumor cell line with acquired MET amplification11,12. Melanoma, renal cell carcinoma, and glioma have all been found to be potentially related to lung cancer. Pathway such as dentatorubropallidoluysian atrophy was predicted here for the first time as being significant pathways in smoking-induced lung cancer. There was a strong coorelation between this pathway and breast cancer prognosis13. In addition, various intracellular signaling pathways and metabolic and other cellular processes were found to be closely related to lung cancer.
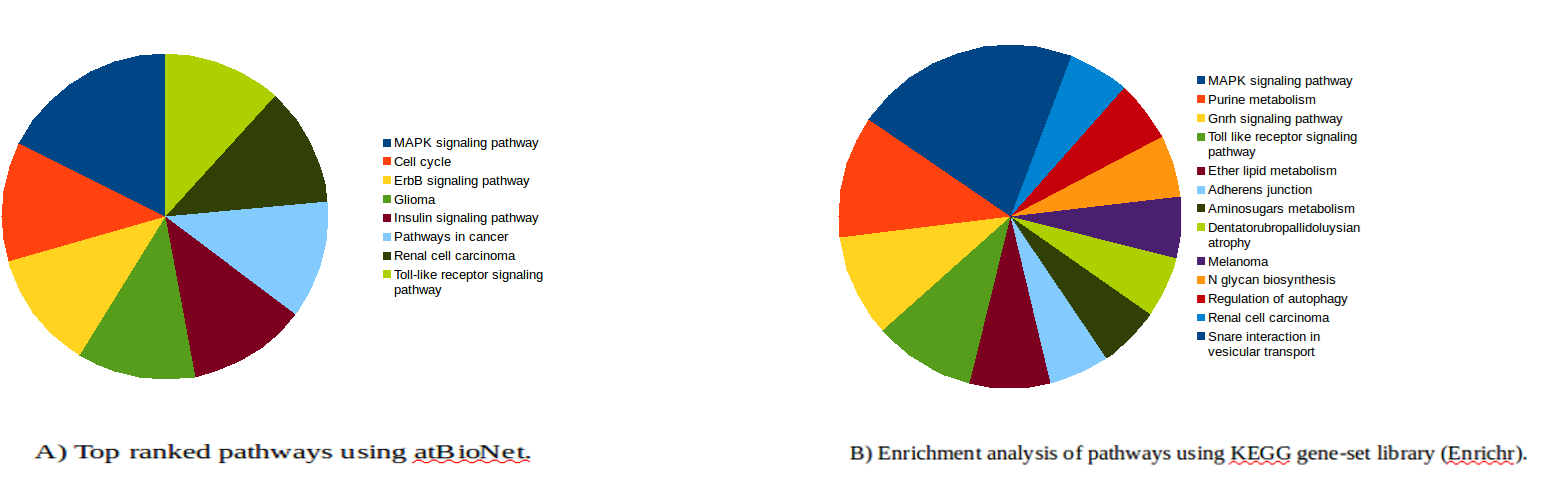
Figure 1
Taken together, these new findings may contribute to the development of ficolin-2 as a novel immunotherapeutic agent that can prevent several important diseases including cancers and infectious diseases.
We found that cyclic AMP-responsive element-binding protein 1(CREB1), nuclear ubiquitous casein and cyclin-dependent kinase substrate 1A, P1 (NUCKS1), Homeobox protein Hox-B4 (HOXB4), and N-myc proto-oncogene protein (MYCN) were the most potential significant transcription factors in lung cancer among smokers (Fig. 2)14-23. Cone-rod homeobox protein (CRX), GA binding protein (GABP), and transcription factor 3 (TCF3) have not been previously implicated in smoking-induced lung cancer. CRX and TCF3 have been detected in other cancers24-26. GABP enables cells to escape apoptosis27.
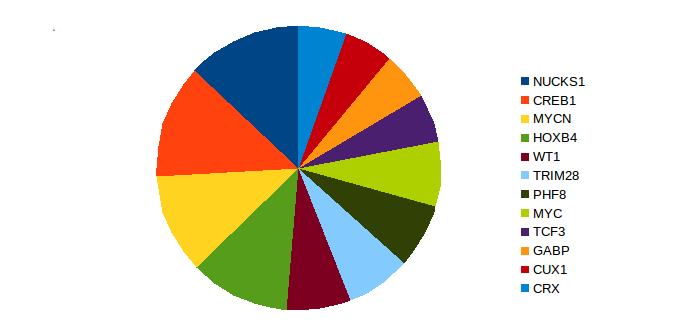
Figure 2
Mitogen-activated protein kinase 1 (MAPK1), insulin-like growth factor 1 receptor (IGFIR), and ribosomal protein S6 kinase alpha-1 (RPS6KA1) were found to be highly related to smoking-induced lung cancer (Fig. 3)28-35. We predicted 5'-AMP-activated protein kinase catalytic subunit alpha-1 (PRKAA1) as a new putative kinase in lung cancer. It can potentially serve as a therapeutic target in chronic myelomonocytic leukemia36.
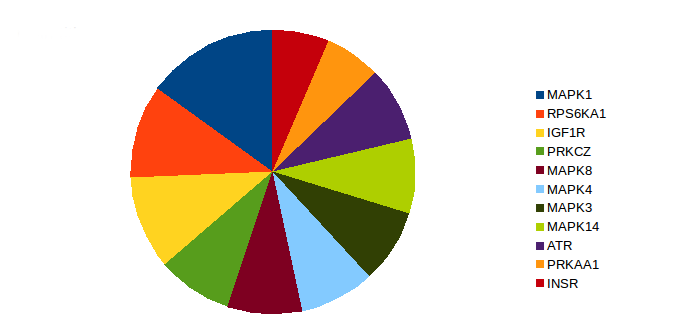
Figure 3
We identified Eight proteins as potential hubs in lung cancer-associated signaling—i.e., Methylphosphate capping enzyme, cyclin-dependent kinase 1, protein kinase C alpha, COP9 signalosome subunit 5, glycogen synthase kinase 3B, breast cancer 1, E1A-binding protein P300, and peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Fig. 4)2,37-48.
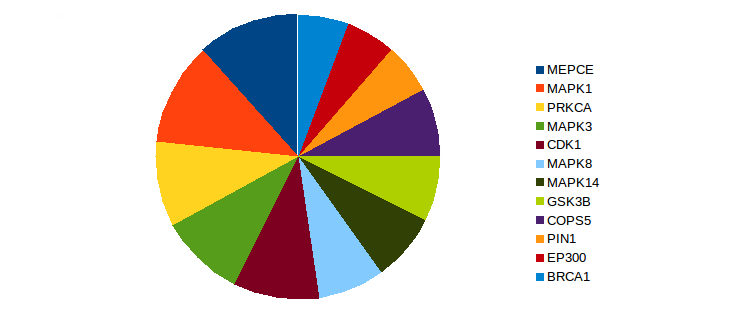
Figure 4
Conclusion
The approach used in this study found well-known key factors which previously reported and validated as drug targets for lung cancer treatment and new potential factors which we claimed. Further study is needed to validate our new potential key regulatory fators in lung cancer among smokers.
References
- Mani KM, Lefebvre C, Wang K, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B- cell lymphomas. Mol Syst Biol. 2008; 7: 1–9.
- Wang YC , Chen BS. A network-based biomarker approach for molecular investigation and diagnosis of lung cancer. BMC Medical Genomics. 2011; 4: 2.
- In silico identification of potential key regulatory factors in smoking-induced lung cancer. BMC Medical Genomics. 2017 Jun 7; 10(1): 40.
- Guo NL, Wan YW. Pathway-based identification of a smoking associated 6-gene signature predictive of lung cancer risk and survival. Artif Intell Med. 2012; 55(2): 97-105.
- Jin Y, Ratnam K, Chuang PY, et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat Med 2012; 18:580-8.
- Lachmann A, Xu H, Krishnan J, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010; 26: 2438–2444.
- Ding YJ, Chen MJ, Liu ZC, et al. atBioNet—an integrated network analysis tool for genomics and biomarker discovery. BMC Genomics. 2012;13, article 325.
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013; 14: 128.
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007; 104(52): 20932–20937.
- Dhillon AS, Hagan S, Rath O, et al. MAP kinase signalling pathways in cancer. Oncogene. 2007; 26: 3279–3290.
- Xing Ke, Meng Wu, Jianfang Lou, et al. Activation of Toll-like receptors signaling in non-small cell lung cancer cell line induced by tumor-associated macrophages. Chin J Cancer Res. 2015; 27(2): 181–189.
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007; 104(52): 20932–20937.
- Giubellino A, Linehan WM, Bottaro DP: Targeting the Met signaling pathway in renal cancer. Expert Rev Anticancer Ther. 2009; 9: 785–793.
- Ma S, Kosorok MR. Detection of gene pathways with predictive power for breast cancer prognosis. BMC Bioinformatics. 2010; 11: 1.
- Xiao X, Li BX, Mitton B, et al. Targeting CREB for cancer therapy: friend or foe. Curr Cancer Drug Targets. 2010; 7: 384–391.
- Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: implications for targeting transcription factors for cancer therapy. Clin Cancer Res. 2009; 15(8): 2583–2587.
- Yan Z, Shah PK, Amin SB, et al. Integrative analysis of gene and miRNA expression profiles with transcription factor-miRNA feed-forward loops identifies regulators in human cancers. Nucleic acids Res. 2012; 40: e135.
- Ryu BJ, Lee H, Kim SH, et al. PF-3758309, p21-activated kinase 4 inhibitor, suppresses migration and invasion of A549 human lung cancer cells via regulation of CREB, NF-kappaB, and beta-catenin signalings. Mol Cell Biochem. 2014; 389: 69–77.
- Sargent LM, Ensell MX, Ostvold AC, et al. Chromosomal changes in high- and low-invasive mouse lung adenocarcinoma cell strains derived from early passage mouse lung adenocarcinoma cell strains. Toxicol Appl Pharmacol. 2008; 233: 81–91.
- Qiu B, Han W, Tergaonkar V. NUCKS: a potential biomarker in cancer and metabolic disease. Clin Sci (Lond). 2015; 128(10): 715-21.
- Lin SH, Wang J, Saintigny P, et al. Genes suppressed by DNA methylation in non-small cell lung cancer reveal the epigenetics of epithelial-mesenchymal transition. BMC Genomics. 2014; 15: 1079.
- Pradhan MP, Desai A, Palakal MJ. Systems biology approach to stage-wise characterization of epigenetic genes in lung adenocarcinoma. BMC Syst Biol. 2013; 7: 141–1.
- Pistoia V, Morandi F, Pezzolo A, et al. MYCN: from oncoprotein to tumor-associated antigen. Front Oncol. 2012; 2: 174.
- Wong AJ, Ruppert JM, Eggleston J, et al. Gene amplification of c-myc and N-myc in small cell carcinoma of the lung. Science. 1986; 233: 461–464.
- Santagata S, Maire CL, Idbaih A, et al. CRX is a diagnostic marker of retinal and pineal lineage tumors. PLoS One. 2009; 4(11): e7932.
- Garrisi VM, Strippoli S, De Summa S, et al. Proteomic Profile and In Silico Analysis in Metastatic Melanoma with and without BRAF Mutation. PLoS ONE. 2014; 9(12): e112025.
- Mo ML, Chen Z, Zhou HM, et al. Detection of E2A-PBX1 fusion transcripts in human non-small-cell lung cancer. J Exp Clin Cancer Res. 2013; 32: 29.
- Bell RJ, Rube HT, Kreig A, et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015; 348: 1036–1039. doi: 10.1126/science.aab0015.
- Buonato JM, Lazzara MJ. ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res. 2014; 74: 309–319. doi: 10.1158/0008-5472.CAN-12-4721.
- Yang L,Su T, Lv D, et al. ERK1/2 mediates lung adenocarcinoma cell proliferation and autophagy induced by apelin-13.Acta Biochim Biophys Sin Shanghai. 2014; 46(2): 100-11.
- Hye-Young Min, Hye Jeong Yun, Ji-Sun Lee, et al. Targeting the insulin-like growth factor receptor and Src signaling network for the treatment of non-small cell lung cancer. Mol Cancer. 2015; 14: 113.
- Kim JS, Kim ES, Liu D, et al. Activation of insulin-like growth factor 1 receptor in patients with non-small cell lung cancer. Oncotarget. 2015; 6(18): 16746–16756.
- Fidler MJ, Shersher DD, Borgia JA, et al. Targeting the insulin-like growth factor receptor pathway in lung cancer: problems and pitfalls. Therapeutic advances in medical oncology. 2012; 4: 51–60.
- Bianconi F, Baldelli E, Ludovini V, et al. Computational model of EGFR and IGF1R pathways in lung cancer: A systems biology approach for translational oncology. Biotechnol Adv. 2012; 30(1): 142–53.
- Slattery ML, Lundgreen A, Herrick JS, et al. Genetic variation in RPS6KA1, RPS6KA2, RPS6KB1, RPS6KB2, and PDK1 and risk of colon or rectal cancer. Mutation research. 2011; 706: 13–20.
- Eisinger-Mathason TS, Andrade J, Lannigan DA. RSK in tumorigenesis: Connections to steroid signaling. Steroids. 2010; 75(3): 191–202.
- Obba S, Hizir Z, Boyer L, et al. The PRKAA1/AMPKα1 pathway triggers autophagy during CSF1-induced human monocyte differentiation and is a potential target in CMML. Autophagy. 2015; 11(7): 1114-29.
- Chang LC, Yu YL, Liu CY, et al. The newly synthesized 2-arylnaphthyridin-4-one, CSC-3436, induces apoptosis of non-small cell lung cancer cells by inhibiting tubulin dynamics and activating CDK1. Cancer Chemother Pharmacol. 2015; 75(6): 1303.
- Abera MB, Kazanietz MG. Protein kinase Cα mediates erlotinib resistance in lung cancer cells. Mol Pharmacol. 2015; 87(5): 832-41.
- Osoegawa A, Yoshino I, Kometani T, et al. Overexpression of Jun activation domain-binding protein 1 in nonsmall cell lung cancer and its significance in p27 expression and clinical features. Cancer. 2006; 107: 154–161.
- Hu MD, Xu JC, Fan Y, et al. Hypoxia-inducible factor 1 promoter-induced JAB1 overexpression enhances chemotherapeutic sensitivity of lung cancer cell line A549 in an anoxic environment. Asian Pac J Cancer Prev. 2012; 13(5): 2115-20.
- Remsing Rix LL, Kuenzi BM, Luo Y, et al. GSK3 alpha and beta are new functionally relevant targets of tivantinib in lung cancer cells. ACS Chem Biol. 2014; 9: 353–8.
- Li Z, Qing Y, Guan W, et al. Predictive value of APE1, BRCA1, ERCC1 and TUBB3 expression in patients with advanced non-small cell lung cancer (NSCLC) receiving first-line platinum-paclitaxel chemotherapy. Cancer Chemother. Pharmacol. 2014; 74: 777–786.
- Rosell R, Skrzypski M, Jassem E, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One. 2007; 2: e1129.
- Cao JX, Lu Y, Qi JJ, et al. MiR-630 inhibits proliferation by targeting CDC7 kinase, but maintains the apoptotic balance by targeting multiple modulators in human lung cancer A549 cells. Cell Death Dis. 2014; 5: e1426.
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics. 2012; 44(10): 1104–1110.
- Yoon HE, Kim SA, Choi HS, et al. Inhibition of Plk1 and Pin1 by 5′-nitro indirubinoxime suppresses human lung cancer cells. Cancer Lett. 2011; 316(1): 97-104.
- Tan X, Zhou F, Wan J, et al. Pin1 expression contributes to lung cancer: Prognosis and carcinogenesis. Cancer Biol Ther. 2010; 9(2): 111-19.
